Open Targets: pre-competitive insights for systematic drug target selection


About Open Targets
Open Targets is a pre-competitive, public-private partnership based at the Wellcome Genome Campus, bridging the gap between academic research and pharmaceutical drug discovery.
Founded in 2014 by the European Bioinformatics Institute, the Wellcome Sanger Institute, and GSK, Open Targets aims to be a world leading centre transforming human target identification and prioritisation by combining groundbreaking experimental and computational approaches to enable systematic selection and validation of targets at scale.
Since Open Targets was founded the partnership has expanded to include additional partners: Sanofi, BMS and Pfizer, who joined at the beginning of 2022.

EMBL-EBI South, the home of Open Targets. Helena Cornu
EMBL-EBI South, the home of Open Targets. Helena Cornu
The problem
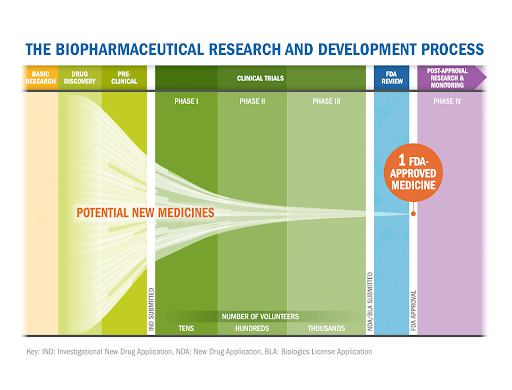
Developing new drugs is a lengthy, costly process, with a low success rate. Many potential therapies fail during clinical trials due to a lack of safety or a lack of efficacy; this indicates that our understanding of the chosen target-disease pair to drug is incomplete or flawed.
However, recent work has shown that therapies supported by human genetic evidence are more likely to progress through the clinical pipeline — up to eight times as likely if the target is supported by Mendelian genetic evidence.
Open Targets’ mission
Open Targets aims to improve the success rate of drug discovery in two ways. First, by enabling drug discovery scientists to explore publicly available data that associates targets and disease with an emphasis on genetic evidence in a seamless and harmonised way, and prioritise relevant targets for their therapeutic area. Second, by filling in the gaps in the existing body of evidence through large-scale, systematic experimental research in neurodegenerative disease biology, oncology, and immunology and inflammation.
Informatics projects
Open Targets’ flagship informatics projects are the Open Targets Platform and Open Targets Genetics.
The Open Targets Platform integrates public data relevant to the association of targets and diseases, and provides additional data and tools to help users prioritise the most interesting targets. Targets covered include protein-coding genes as well as other molecules such as non-coding RNAs. The Platform ingests data to associate targets with diseases and phenotypes from gold-standard public repositories as well as including data generated within the consortium.

Users exploring the Open Targets informatics tools at an Open Targets integration day.
Users exploring the Open Targets informatics tools at an Open Targets integration day.
Open Targets Platform landing page
Open Targets Platform landing page
The Open Targets Platform
The Platform is the partnership’s flagship product, developed together with scientists from the partner institutions through a user experience design process.
It is an open source project which is constantly updated and improved. The team publishes five releases a year, integrating the latest data and creating new functionalities to maximise the use of the data.
The Open Targets Genetics portal
One of the Platform’s data sources is Open Targets Genetics. This portal provides causal target assignments underlying each association for Genome Wide Association Studies. Open Targets recently developed a machine learning model to assign each association at a locus to the most likely causal gene based on integrated genetics and functional genomics data.
Open Targets also supports data resources such as the eQTL Catalogue, and has an ecosystem of projects in the area of bioinformatics, some of which feed into the Open Targets Platform.
Open Targets Genetics landing page
Open Targets Genetics landing page
Experimental projects
Open Targets’ portfolio of experimental projects, designed collaboratively by our academic and pharmaceutical partners, provides new information in our key therapy areas to enable target identification and prioritisation.
The projects focus on human, physiologically relevant systems, to generate new data that either supports causal associations between targets and diseases, or provides a foundational understanding of disease biology in oncology, neurodegeneration, and immunology and inflammation.
The consortium has already made significant contributions to understanding the relationship between targets and diseases. In 2019, a project led by Matthew Garnett and Fiona Behan as part of Open Targets’ Project SCORE performed whole genome CRISPR-Cas9 screening in over 300 cell lines from 30 cancer types to identify new cancer targets. This work contributed to the Cancer Dependency Map project, a collaboration between the Wellcome Sanger Institute and the Broad Institute.
The team identified and verified Werner syndrome ATP-dependent helicase as a synthetic lethal target in tumours with microsatellite instability. Their data was integrated into the Open Targets Platform, to inform target prioritisation in oncology.



Open targets team at the Hinxton Hall Conference Centre, Wellcome Genome Campus
Open targets team at the Hinxton Hall Conference Centre, Wellcome Genome Campus

Cell culture bottles of the CAL-120 breast cancer cell line, prepared for a CRISPR screen as part of an Open Targets oncology project. Jessica Cantwell]
Cell culture bottles of the CAL-120 breast cancer cell line, prepared for a CRISPR screen as part of an Open Targets oncology project. Jessica Cantwell]

Cancer cells. Credit: National Cancer institute
Cancer cells. Credit: National Cancer institute
What’s next?
With an ambitious vision for the future, we are developing approaches to enable drug discovery scientists to systematically construct and assess therapeutic hypotheses.
We want to empower scientists to think of their chosen target in the context of its interacting network of molecules, and how this context changes throughout disease progression. For example, Open Targets is working to generate and integrate new datasets to provide granular data on the cell type, state, and tissue where we want therapies to work, as well as the diseases, subtypes, stages and biomarkers.
By bringing therapeutic hypothesis construction to the fore, we hope to prompt ideas and discussion to guide how future versions of our informatics ecosystem and wider research programme can evolve.
Know more about Open Targets on their website: https://www.opentargets.org/